These functions can be used for generating random MIC values and disk diffusion diameters, for AMR data analysis practice. By providing a microorganism and antimicrobial drug, the generated results will reflect reality as much as possible.
Usage
random_mic(size = NULL, mo = NULL, ab = NULL, skew = "right",
severity = 1, ...)
random_disk(size = NULL, mo = NULL, ab = NULL, skew = "left",
severity = 1, ...)
random_sir(size = NULL, prob_SIR = c(0.33, 0.33, 0.33), ...)Arguments
- size
Desired size of the returned vector. If used in a data.frame call or
dplyrverb, will get the current (group) size if left blank.- mo
Any character that can be coerced to a valid microorganism code with
as.mo(). Can be the same length assize.- ab
Any character that can be coerced to a valid antimicrobial drug code with
as.ab().- skew
Direction of skew for MIC or disk values, either
"right"or"left". A left-skewed distribution has the majority of the data on the right.- severity
Skew severity; higher values will increase the skewedness. Default is
2; use0to prevent skewedness.- ...
Ignored, only in place to allow future extensions.
- prob_SIR
A vector of length 3: the probabilities for "S" (1st value), "I" (2nd value) and "R" (3rd value).
Details
Internally, MIC and disk zone values are sampled based on clinical breakpoints defined in the clinical_breakpoints data set. To create specific generated values per bug or drug, set the mo and/or ab argument. The MICs are sampled on a log2 scale and disks linearly, using weighted probabilities. The weights are based on the skew and severity arguments:
skew = "right"places more emphasis on lower MIC or higher disk values.skew = "left"places more emphasis on higher MIC or lower disk values.severitycontrols the exponential bias applied.
Examples
random_mic(25)
#> Class 'mic'
#> [1] 0.125 <=0.0001 0.004 0.004 0.5 0.032 1 0.008
#> [9] 128 0.0002 0.002 0.002 32 0.032 0.0005 0.0002
#> [17] 0.008 0.001 0.0002 <=0.0001 0.064 0.016 0.008 4
#> [25] 1
random_disk(25)
#> Class 'disk'
#> [1] 50 21 48 47 46 38 45 47 24 28 45 43 19 23 44 19 31 22 40 39 39 36 35 14 34
random_sir(25)
#> Class 'sir'
#> [1] I S S R I S R I R I S S S R R R I I R R I S R R S
# add more skewedness, make more realistic by setting a bug and/or drug:
disks <- random_disk(100, severity = 2, mo = "Escherichia coli", ab = "CIP")
plot(disks)
 # `plot()` and `ggplot2::autoplot()` allow for coloured bars if `mo` and `ab` are set
plot(disks, mo = "Escherichia coli", ab = "CIP", guideline = "CLSI 2025")
# `plot()` and `ggplot2::autoplot()` allow for coloured bars if `mo` and `ab` are set
plot(disks, mo = "Escherichia coli", ab = "CIP", guideline = "CLSI 2025")
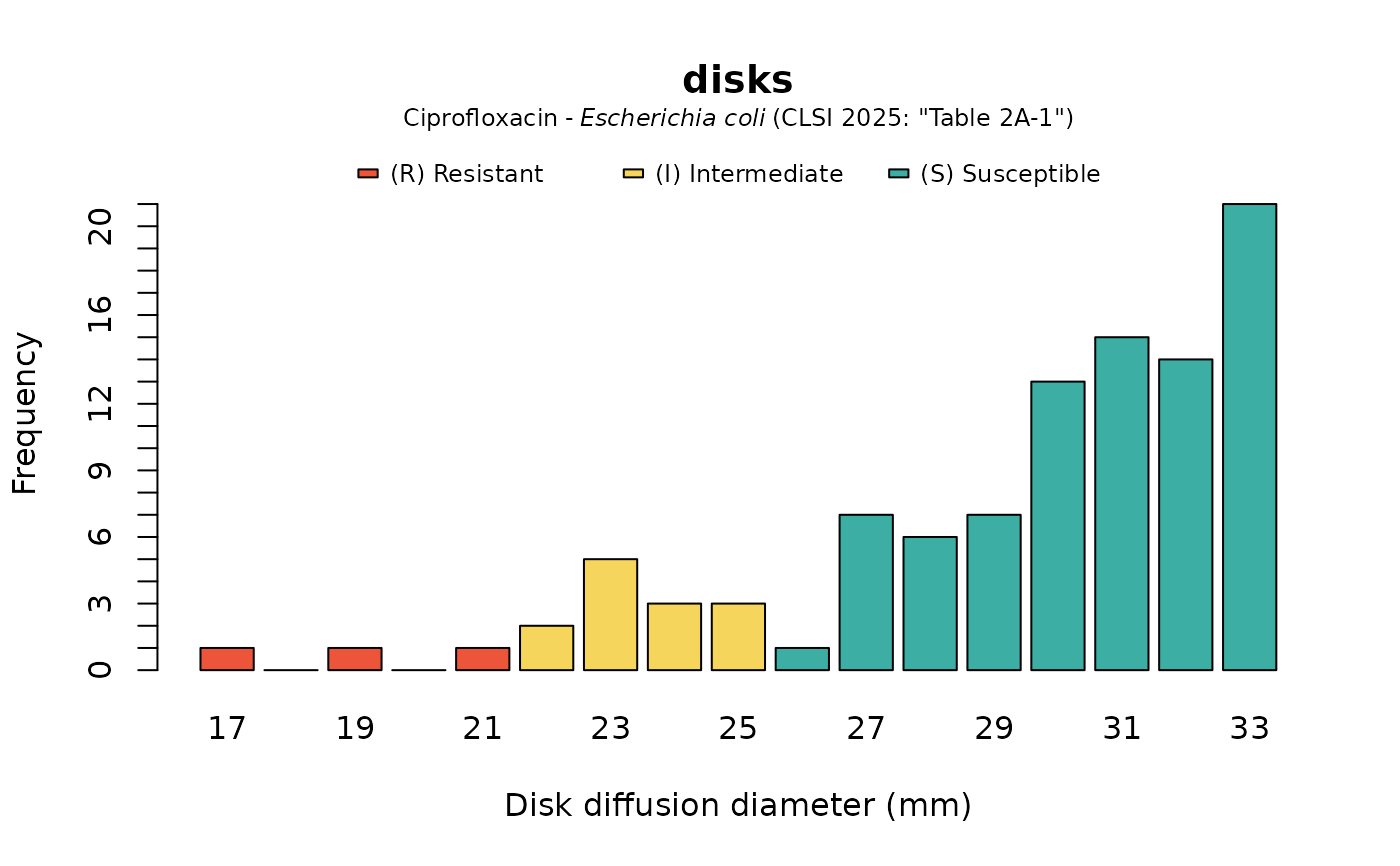 # \donttest{
random_mic(25, "Klebsiella pneumoniae") # range 0.0625-64
#> Class 'mic'
#> [1] 0.0001 0.0002 0.016 0.5 0.008 >=8 0.125 0.0002 0.002 0.0005
#> [11] 1 0.0001 0.25 0.0002 0.25 2 0.0005 0.064 0.5 0.004
#> [21] 0.001 0.064 0.032 0.0005 0.125
random_mic(25, "Klebsiella pneumoniae", "meropenem") # range 0.0625-16
#> Class 'mic'
#> [1] <=2 <=2 4 <=2 <=2 4 <=2 4 <=2 4 <=2 <=2 <=2 <=2 <=2 <=2 4 4 <=2
#> [20] 4 <=2 <=2 <=2 <=2 <=2
random_mic(25, "Streptococcus pneumoniae", "meropenem") # range 0.0625-4
#> Class 'mic'
#> [1] 2 <=0.064 1 <=0.064 <=0.064 2 <=0.064 0.25 0.25
#> [10] 2 <=0.064 <=0.064 <=0.064 <=0.064 0.25 0.5 0.5 0.25
#> [19] 0.125 0.125 4 0.25 1 <=0.064 <=0.064
random_disk(25, "Klebsiella pneumoniae") # range 8-50
#> Class 'disk'
#> [1] 14 19 19 33 33 26 16 27 29 33 24 17 17 15 16 26 26 14 17 29 25 25 24 32 32
random_disk(25, "Klebsiella pneumoniae", "ampicillin") # range 11-17
#> Class 'disk'
#> [1] 22 15 21 11 19 20 11 14 12 12 17 16 12 10 15 22 19 21 19 17 18 19 21 17 21
random_disk(25, "Streptococcus pneumoniae", "ampicillin") # range 12-27
#> Class 'disk'
#> [1] 21 25 16 29 27 24 31 22 35 28 26 32 24 18 35 33 24 28 33 25 35 25 32 24 26
# }
# \donttest{
random_mic(25, "Klebsiella pneumoniae") # range 0.0625-64
#> Class 'mic'
#> [1] 0.0001 0.0002 0.016 0.5 0.008 >=8 0.125 0.0002 0.002 0.0005
#> [11] 1 0.0001 0.25 0.0002 0.25 2 0.0005 0.064 0.5 0.004
#> [21] 0.001 0.064 0.032 0.0005 0.125
random_mic(25, "Klebsiella pneumoniae", "meropenem") # range 0.0625-16
#> Class 'mic'
#> [1] <=2 <=2 4 <=2 <=2 4 <=2 4 <=2 4 <=2 <=2 <=2 <=2 <=2 <=2 4 4 <=2
#> [20] 4 <=2 <=2 <=2 <=2 <=2
random_mic(25, "Streptococcus pneumoniae", "meropenem") # range 0.0625-4
#> Class 'mic'
#> [1] 2 <=0.064 1 <=0.064 <=0.064 2 <=0.064 0.25 0.25
#> [10] 2 <=0.064 <=0.064 <=0.064 <=0.064 0.25 0.5 0.5 0.25
#> [19] 0.125 0.125 4 0.25 1 <=0.064 <=0.064
random_disk(25, "Klebsiella pneumoniae") # range 8-50
#> Class 'disk'
#> [1] 14 19 19 33 33 26 16 27 29 33 24 17 17 15 16 26 26 14 17 29 25 25 24 32 32
random_disk(25, "Klebsiella pneumoniae", "ampicillin") # range 11-17
#> Class 'disk'
#> [1] 22 15 21 11 19 20 11 14 12 12 17 16 12 10 15 22 19 21 19 17 18 19 21 17 21
random_disk(25, "Streptococcus pneumoniae", "ampicillin") # range 12-27
#> Class 'disk'
#> [1] 21 25 16 29 27 24 31 22 35 28 26 32 24 18 35 33 24 28 33 25 35 25 32 24 26
# }